OrgMassSpecR
OrgMassSpecR is an extension for the R statistical computing language.
It contains
functions to assist with organic/biological mass spectrometry data analysis. Mass spectral libraries are
available
as companion packages.
This project is under continuous development. The stable version of OrgMassSpecR is on CRAN.
The development version of OrgMassSpecR is on GitHub.
The mass spectral library packages and stand-alone files (PDF reports and MSP files) are available on the GitHub
Mass Spectral Libraries page.
OrgMassSpecR examples are in the package
vignette.
Functions
General
MolecularWeight: Calculate the molecular weight of an organic molecule.
MonoisotopicMass: Calculate the monoisotopic mass or monoisotopic m/z value of an organic
molecule.
IsotopicDistribution: Simulate the isotopic distribution of an organic molecule.
ListFormula: Convert a character string representing an elemental formula to a list
representing
the elemental formula.
DrawChromatogram: Plot a chromatogram, color the area under specified peak(s), and calculate
the peak area(s).
SpectrumSimilarity: Generate a head-to-tail plot of two mass spectra and calculate a
similarity
score.
ConvertConcentration: Change the unit basis for a sample concentration, such as ng/g wet
weight
to ng/g dry weight, or pg/g lipid weight to pg/g wet weight.
RetentionIndex: Calculate the the non-isothermal gas chromatographic retention index of a
target
compound.
DeadVolume: Calculate the internal volume of a defined length of tubing.
FlowTime: Calculate the time required for a liquid to flow through a defined length of
tubing.
Spectral
Libraries
LibraryReport: Generate a PDF report of an OrgMassSpecR mass spectral library, or view
spectra
within R. Each spectral library package has its own LibraryReport function.
ReadMspDirectory: Reads in all .msp files within a directory and makes a single concatenated
data frame of m/z values and intensities.
ReadMspFile: Reads in a .msp file and makes a data frame of m/z values and intensities.
WriteMspFile: Writes a single .msp file from a data frame of m/z values and intensities of
multiple
mass spectra.
Proteins/Peptides
Digest: Cleave an amino acid sequence (a protein or peptide) according to enzyme specific rules
and calculate the precursor ion m/z values.
FragmentPeptide: Determine the b- and y-ions or c- and z-ions produced by the fragmentation of
a peptide by tandem mass spectrometry.
ConvertPeptide: Convert single amino acid codes to an elemental formula or three letter codes.
PeptideSpectrum: Plot a peptide fragmentation mass spectrum, with the b- and y-ions or c- and
z-ions identified.
IsotopicDistributionN: Simulate the isotopic distribution of a peptide with varying amounts of
nitrogen-15 incorporation.
IsotopicDistributionHDX: Simulate the isotopic distribution of a peptide undergoing
hydrogen-deuterium
exchange.
ExchangeableAmides: Determine the number of backbone amide hydrogens given a protein/peptide
sequence.
Used in hydrogen-deuterium exchange experiments.
Examples
Halogenated Isotopic Distributions
The characteristic isotopic distributions of bromine and chlorine atoms can aid the interpretation of mass
spectra.
Bromine Isotopic Distributions (PDF)
Chlorine Isotopic Distributions (PDF)
Bromine + Chlorine Isotopic
Distributions (PDF)
Example Graphics
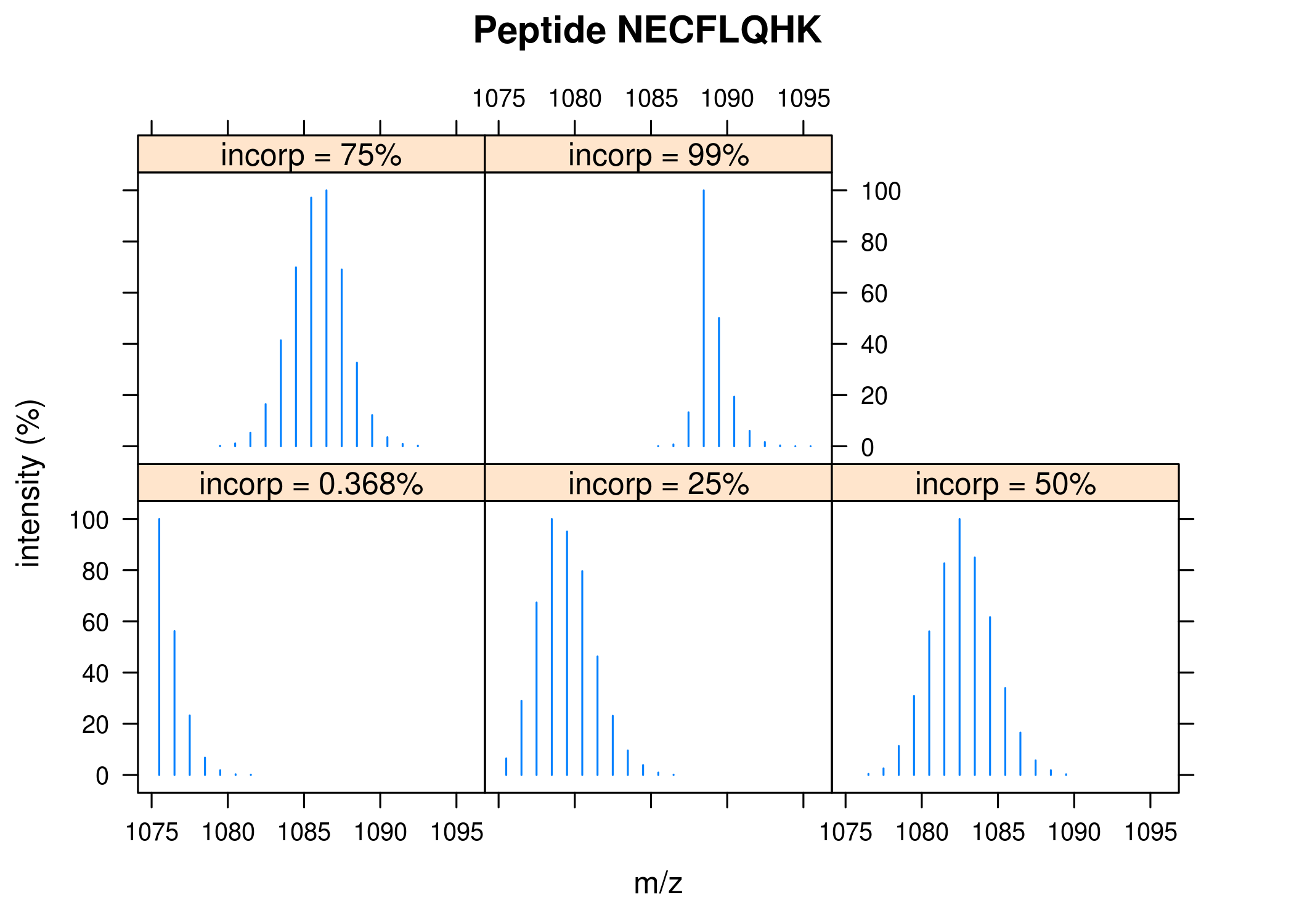
Simulated isotopic distributions of a peptide with varying nitrogen-15 incorporation. Function
IsotopicDistributionN.
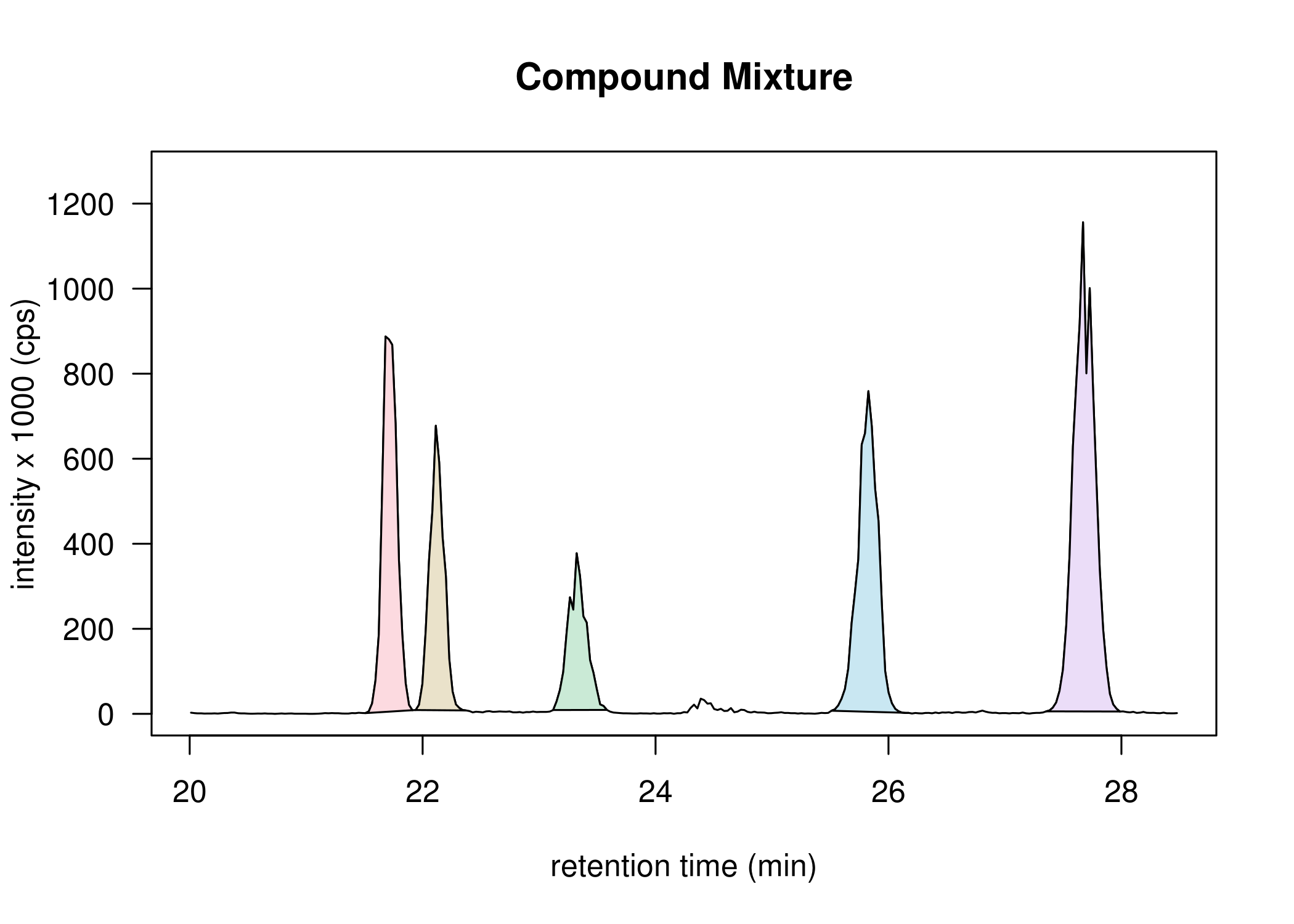
Highlighted peaks. Function DrawChromatogram.
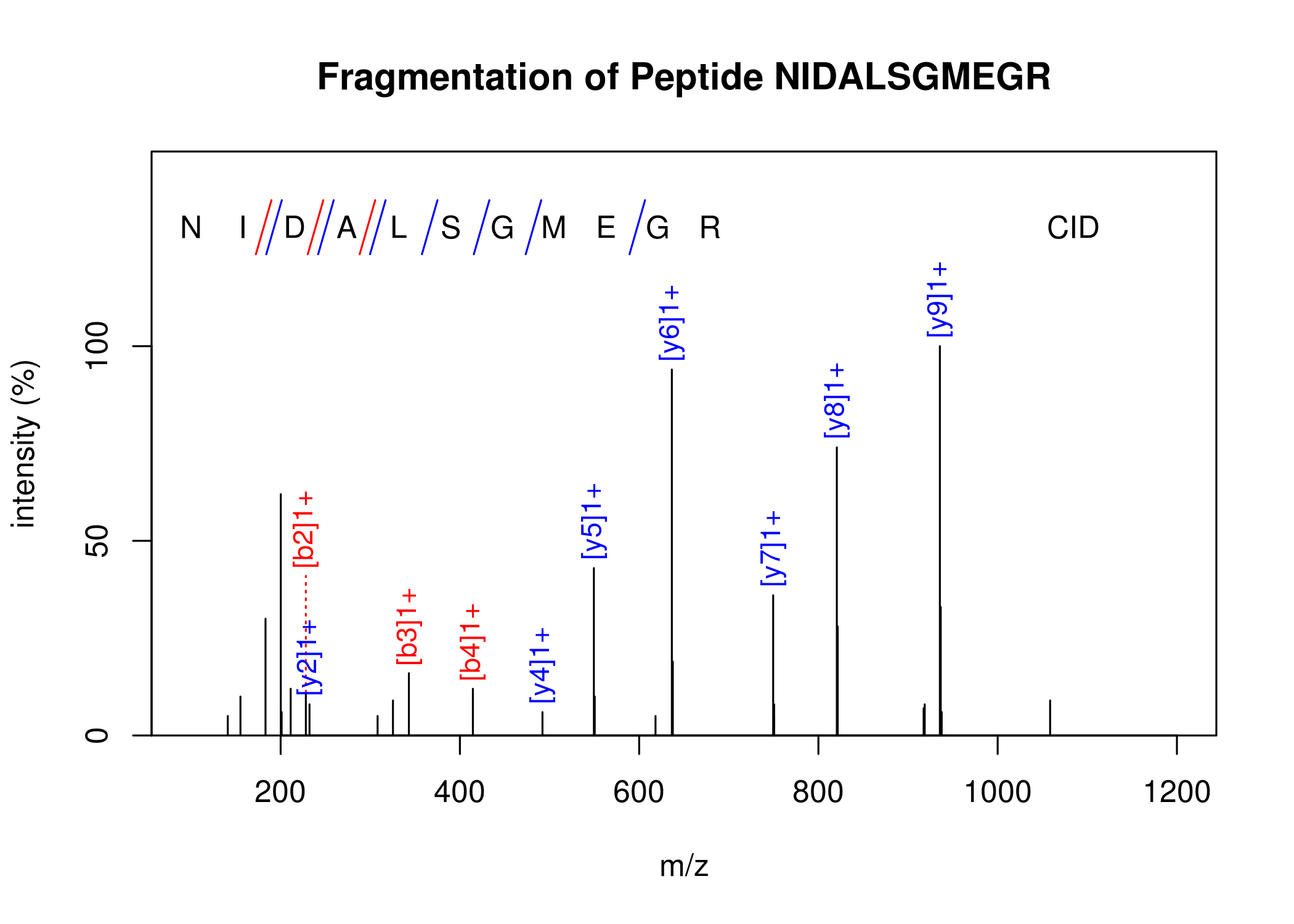
Peptide fragmentation mass spectrum. Function PeptideSpectrum.
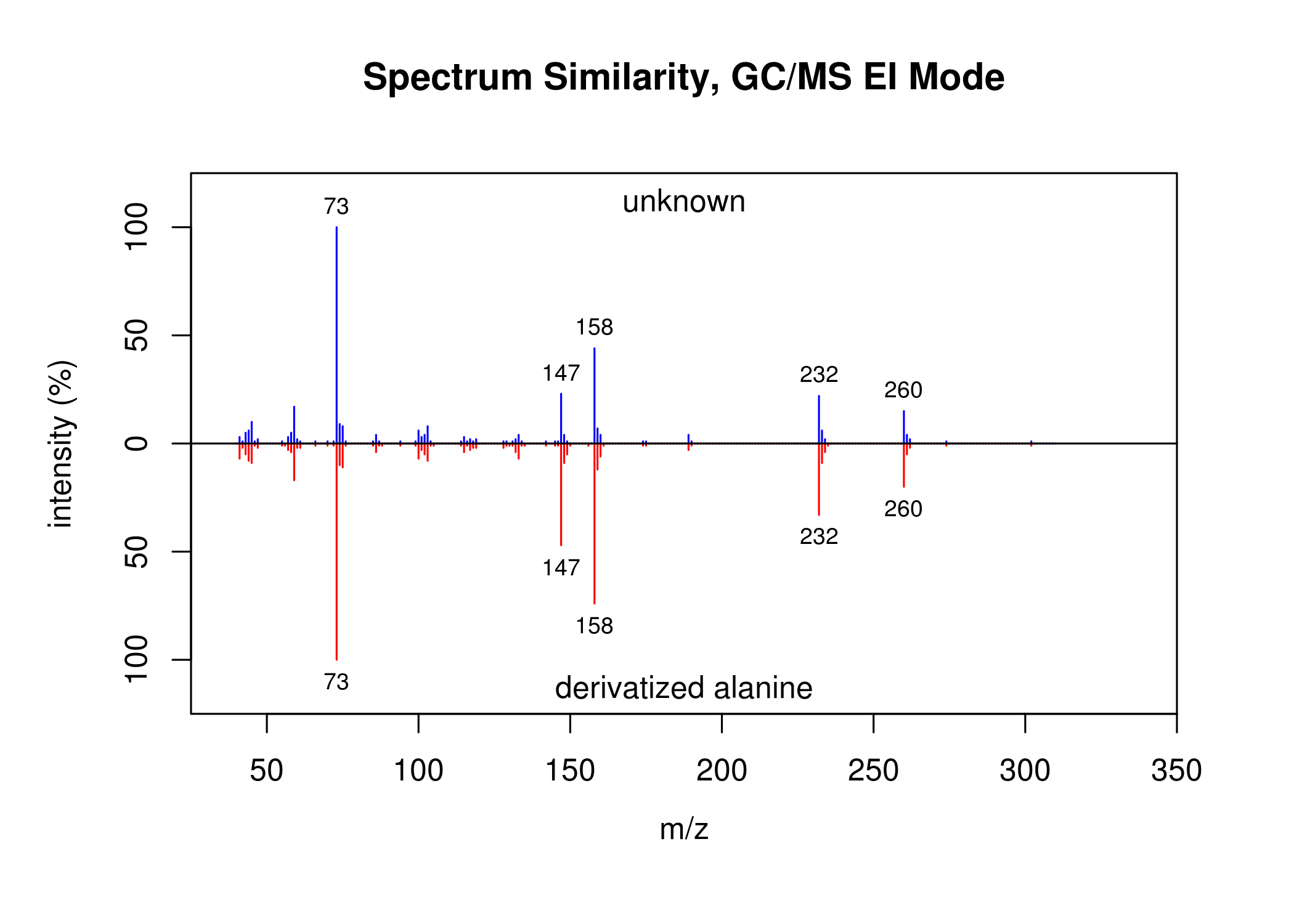
Spectrum similarity. Function SpectrumSimilarity.
References
Introductory
Manuscripts
Mass Spectral Libraries
Hoh E, Dodder NG, Lehotay SJ, Pangallo KC, Reddy CM, Maruya KA. Nontargeted Comprehensive Two-Dimensional Gas
Chromatography/Time-of-Flight
Mass Spectrometry Method and Software for Inventorying Persistent and Bioaccumulative Contaminants in Marine
Environments.
Environ Sci Technol. 2012;46: 8001-8008. [Link]
Calculation of MRM Transitions and Isotopic Distributions of Nitrogen-15 Labeled Peptides
Liao W-L, Heo G-Y, Dodder NG, Pikuleva IA, Turko IV. Optimizing the Conditions of a Multiple Reaction
Monitoring
Assay for Membrane Proteins: Quantification of Cytochrome P450 11A1 and Adrenodoxin Reductase in Bovine Adrenal
Cortex and Retina. Anal Chem. 2010;82: 5760-5767. [Link]
Manuscripts Using or Referring to OrgMassSpecR
1.
Samokhin A, Khrisanfov M. High-Throughput Mass Spectral
Library Searching of Small Molecules in R with NIST MSPepSearch. J Am Soc Mass Spectrom. 2026;37: 264-269.
doi:
10.1021/jasms.5c00322
2.
Ivanova A, Tang W, Simon C, Dührkop K, Böcker S,
Gleixner G. Enhancing Chimeric Fragmentation Spectra Deconvolution Using Direct Infusion-Tandem Mass
Spectrometry
Across High-Resolution Mass Spectrometric Platforms. Rapid Communications in Mass Spectrometry. 2026;40:
e10170.
doi:
10.1002/rcm.10170
3.
Willis B, Whitwell HJ. Protein digestion, peptide mass and peptide
fragmentation with MZCal: A user-friendly phone-compatible application. Journal of Proteomics. 2025;318:
105456. doi:
10.1016/j.jprot.2025.105456
4.
Tammen H, Pich A, Hess R, Lechowicz U, Janciauskiene S, Chorostowska J.
Quantitative mass spectrometric analysis of C-terminal 36 amino acid peptides of alpha-1 antitrypsin in plasma
using survey spectra. Methods. 2025;240: 7-13. doi:
10.1016/j.ymeth.2025.04.004
5.
Hernández-Lao T, Rodríguez-Pérez R, Labella-Ortega M, Muñoz Triviño M,
Pedrosa M, Rey M-D, et al. Proteomic identification of allergenic proteins in holm oak (
Quercus ilex)
seeds. Food Chemistry. 2025;464: 141667. doi:
10.1016/j.foodchem.2024.141667
6.
Gao M, Hu W, Meng D, Yao P, Yang S, Tong Y, et al. Jolkinolide B Activates
Mitophagy to Exhibit Antipancreatic Cancer Activity and Alleviate Cognitive Deficits in Alzheimer's Disease.
Molecular & Cellular Proteomics. 2025;24: 101060. doi:
10.1016/j.mcpro.2025.101060
7.
Cloteau C, Delcourt V, Loup B, Chabot B, Pescher M, Susdorf E, et al.
Identification of Candidate Biomarkers Detected in the Urine of Racehorses After Anabolic Agent
Administration: Use of Orthogonal Methods for Structural Elucidation. Drug Testing and Analysis. 2025;17:
2411-2420. doi:
10.1002/dta.3951
8.
Yan Y, Hemmler D, Schmitt-Kopplin P. Discovery of Glycation Products:
Unraveling the Unknown Glycation Space Using a Mass Spectral Library from In Vitro Model Systems. Anal Chem.
2024;96: 3569-3577. doi:
10.1021/acs.analchem.3c05540
9.
Talavera Andújar B, Mary A, Venegas C, Cheng T, Zaslavsky L, Bolton EE, et
al. Can Small Molecules Provide Clues on Disease Progression in Cerebrospinal Fluid from Mild Cognitive
Impairment and Alzheimer's Disease Patients? Environ Sci Technol. 2024;58: 4181-4192. doi:
10.1021/acs.est.3c10490
10.
Fiala J, Schuster D, Ollivier S, Pengelley S, Lubeck M, Busch F, et al.
Protein-Centric Analysis of Personalized Antibody Repertoires Using LC-MS-Based Fab-Profiling on a timsTOF. J
Am Soc Mass Spectrom. 2024;35: 1292-1300. doi:
10.1021/jasms.4c00076
11.
Albergamo V, Wohlleben W, Plata DL. Tracking Dynamic Chemical Reactivity
Networks with High-Resolution Mass Spectrometry: A Case of Microplastic-Derived Dissolved Organic Carbon.
Environ Sci Technol. 2024;58: 4314-4325. doi:
10.1021/acs.est.3c08134
12.
Kacen A, Javitt A, Kramer MP, Morgenstern D, Tsaban T, Shmueli MD, et al.
Post-translational modifications reshape the antigenic landscape of the MHC I immunopeptidome in tumors. Nat
Biotechnol. 2023;41: 239-251. doi:
10.1038/s41587-022-01464-2
13.
Codrean S, Kruit B, Meekel N, Vughs D, Béen F. Predicting the Diagnostic
Information of Tandem Mass Spectra of Environmentally Relevant Compounds Using Machine Learning. Anal Chem.
2023;95: 15810-15817. doi:
10.1021/acs.analchem.3c03470
14.
Debnath T, Nakamoto T. Extraction of sensing data for desired scent
impressions using mass spectra of odorant molecules. Sci Rep. 2022;12: 16297. doi:
10.1038/s41598-022-20388-0
15.
Ulanga U, Russell M, Patassini S, Brazzatti J, Graham C, Whetton AD, et al.
Generation of a mouse SWATH-MS spectral library to quantify 10148 proteins involved in cell reprogramming. Sci
Data. 2021;8: 118. doi:
10.1038/s41597-021-00896-w
16.
Sisco E, Moorthy AS, Watt LM. Creation and Release of an Updated NIST
DART-MS Forensics Database. J Am Soc Mass Spectrom. 2021;32: 685-689. doi:
10.1021/jasms.0c00416
17.
Meekel N, Vughs D, Béen F, Brunner AM. Online Prioritization of Toxic
Compounds in Water Samples through Intelligent HRMS Data Acquisition. Anal Chem. 2021;93: 5071-5080. doi:
10.1021/acs.analchem.0c04473
18.
Klünemann M, Andrejev S, Blasche S, Mateus A, Phapale P, Devendran S, et
al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature. 2021; 1-6. doi:
10.1038/s41586-021-03891-8
19.
Huber C, Müller E, Schulze T, Brack W, Krauss M. Improving the Screening
Analysis of Pesticide Metabolites in Human Biomonitoring by Combining High-Throughput In Vitro Incubation and
Automated LC-HRMS Data Processing. Anal Chem. 2021;93: 9149-9157. doi:
10.1021/acs.analchem.1c00972
20.
Guo G, Papanicolaou M, Demarais NJ, Wang Z, Schey KL, Timpson P, et al.
Automated annotation and visualisation of high-resolution spatial proteomic mass spectrometry imaging data
using HIT-MAP. Nat Commun. 2021;12: 3241. doi:
10.1038/s41467-021-23461-w
21.
Xue Y, Vughs D, Hater W, Huiting H, Vanoppen M, Cornelissen E, et al.
Liquid Chromatography-High-Resolution Mass Spectrometry-Based Target and Nontarget Screening Methods to
Characterize Film-Forming Amine-Treated Steam-Water Systems. Ind Eng Chem Res. 2020;59: 22301-22309. doi:
10.1021/acs.iecr.0c05051
22.
Ravenhill BJ, Soday L, Houghton J, Antrobus R, Weekes MP. Comprehensive
cell surface proteomics defines markers of classical, intermediate and non-classical monocytes. Scientific
Reports. 2020;10: 1-11. doi:
10.1038/s41598-020-61356-w
23.
Keating MF, Zhang J, Feider CL, Retailleau S, Reid R, Antaris A, et al.
Integrating the MasSpec Pen to the da Vinci Surgical System for In Vivo Tissue Analysis during a Robotic
Assisted Porcine Surgery. Anal Chem. 2020 [cited 21 Aug 2020]. doi:
10.1021/acs.analchem.0c02037
24.
Stanstrup J, Broeckling CD, Helmus R, Hoffmann N, Mathé E, Naake T, et al.
The metaRbolomics Toolbox in Bioconductor and beyond. Metabolites. 2019;9. doi:
10.3390/metabo9100200
25.
Merkley ED, Burnum-Johnson KE, Anderson LN, Jenson SC, Wahl KL. Uniformly
15N-Labeled Recombinant Ricin A-Chain as an Internal Retention Time Standard for Increased Confidence in
Forensic Identification of Ricin by Untargeted Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal
Chem. 2019;91: 13372-13376. doi:
10.1021/acs.analchem.9b03389
26.
Djoumbou-Feunang Y, Pon A, Karu N, Zheng J, Li C, Arndt D, et al. CFM-ID
3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites. 2019;9: 72. doi:
10.3390/metabo9040072
27.
Dennis KK, Uppal K, Liu KH, Ma C, Liang B, Go Y-M, et al. Phytochelatin
database: a resource for phytochelatin complexes of nutritional and environmental metals. Database (Oxford).
2019;2019. doi:
10.1093/database/baz083
28.
Alygizakis NA, Gago-Ferrero P, Hollender J, Thomaidis NS. Untargeted
time-pattern analysis of LC-HRMS data to detect spills and compounds with high fluctuation in influent
wastewater. Journal of Hazardous Materials. 2019;361: 19-29. doi:
10.1016/j.jhazmat.2018.08.073
29.
Albergamo V, Escher BI, Schymanski EL, Helmus R, Dingemans MML, Cornelissen
ER, et al. Evaluation of reverse osmosis drinking water treatment of riverbank filtrate using bioanalytical
tools and non-target screening. Environ Sci: Water Res Technol. 2019;6: 103-116. doi:
10.1039/C9EW00741E
30.
Issa SMA, Vitiazeva V, Hayes CA, Karlsson NG. Higher Energy Collisional
Dissociation Mass Spectrometry of Sulfated O-Linked Oligosaccharides. J Proteome Res. 2018;17: 3259-3267.
doi:
10.1021/acs.jproteome.8b00376
31.
Schollée JE, Schymanski EL, Stravs MA, Gulde R, Thomaidis NS, Hollender J.
Similarity of High-Resolution Tandem Mass Spectrometry Spectra of Structurally Related Micropollutants and
Transformation Products. J Am Soc Mass Spectrom. 2017;28: 2692-2704. doi:
10.1021/jasms.8b05447
32.
Kries H, Kellner F, Kamileen MO, O'Connor SE. Inverted stereocontrol of
iridoid synthase in snapdragon. J Biol Chem. 2017; jbc.M117.800979. doi:
10.1074/jbc.M117.800979
33.
Mackintosh SA, Dodder NG, Shaul NJ, Aluwihare LI, Maruya KA, Chivers SJ, et
al. Newly Identified DDT-Related Compounds Accumulating in Southern California Bottlenose Dolphins. Environ
Sci Technol. 2016;50: 12129-12137. doi:
10.1021/acs.est.6b03150
34.
Shaul NJ, Dodder NG, Aluwihare LI, Mackintosh SA, Maruya KA, Chivers SJ, et
al. Nontargeted biomonitoring of halogenated organic compounds in two ecotypes of bottlenose dolphins
(Tursiops truncatus) from the Southern California Bight. Environ Sci Technol. 2015;49: 1328-1338. doi:
10.1021/es505156q
35.
Scott KB, Turko IV, Phinney KW. Quantitative Performance of Internal
Standard Platforms for Absolute Protein Quantification Using Multiple Reaction Monitoring-Mass Spectrometry.
Anal Chem. 2015 [cited 10 Apr 2015]. doi:
10.1021/acs.analchem.5b00331
36.
Schollée JE, Schymanski EL, Avak SE, Loos M, Hollender J. Prioritizing
Unknown Transformation Products from Biologically-Treated Wastewater Using High-Resolution Mass Spectrometry,
Multivariate Statistics, and Metabolic Logic. Anal Chem. 2015;87: 12121-12129. doi:
10.1021/acs.analchem.5b02905
37.
Rodriguez-Garcia M, Surman AJ, Cooper GJT, Suárez-Marina I, Hosni Z, Lee
MP, et al. Formation of oligopeptides in high yield under simple programmable conditions. Nature
Communications. 2015;6: 8385. doi:
10.1038/ncomms9385
38.
Gago-Ferrero P, Schymanski EL, Bletsou AA, Aalizadeh R, Hollender J,
Thomaidis NS. Extended Suspect and Non-Target Strategies to Characterize Emerging Polar Organic Contaminants
in Raw Wastewater with LC-HRMS/MS. Environ Sci Technol. 2015;49: 12333-12341. doi:
10.1021/acs.est.5b03454
39.
ElBashir R, Vanselow JT, Kraus A, Janzen CJ, Siegel TN, Schlosser A.
Fragment Ion Patchwork Quantification for Measuring Site-Specific Acetylation Degrees. Anal Chem. 2015;87:
9939-9945. doi:
10.1021/acs.analchem.5b02517
40.
Anderson KW, Chen J, Wang M, Mast N, Pikuleva IA, Turko IV. Quantification
of Histone Deacetylase Isoforms in Human Frontal Cortex, Human Retina, and Mouse Brain. PLOS ONE. 2015;10:
e0126592. doi:
10.1371/journal.pone.0126592
41.
Gatto L, Christoforou A. Using R and Bioconductor for proteomics data
analysis. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2014;1844: 42-51. doi:
10.1016/j.bbapap.2013.04.032
42.
Dittwald P, Vu TN, Harris GA, Caprioli RM, Van de Plas R, Laukens K, et al.
Towards automated discrimination of lipids versus peptides from full scan mass spectra. EuPA Open Proteomics.
2014;4: 87-100. doi:
10.1016/j.euprot.2014.05.002
43.
Campbell MP, Nguyen-Khuong T, Hayes CA, Flowers SA, Alagesan K, Kolarich D,
et al. Validation of the curation pipeline of UniCarb-DB: Building a global glycan reference MS/MS repository.
Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2014;1844: 108-116. doi:
10.1016/j.bbapap.2013.04.018
44.
Zushi Y, Hashimoto S, Fushimi A, Takazawa Y, Tanabe K, Shibata Y. Rapid
automatic identification and quantification of compounds in complex matrices using comprehensive
two-dimensional gas chromatography coupled to high resolution time-of-flight mass spectrometry with a peak
sentinel tool. Analytica Chimica Acta. 2013;778: 54-62. doi:
10.1016/j.aca.2013.03.049
45.
Tang Z, Wu M, Li Y, Zheng X, Liu H, Cheng X, et al. Absolute quantification
of NAD(P)H:quinone oxidoreductase 1 in human tumor cell lines and tissues by liquid chromatography-mass
spectrometry/mass spectrometry using both isotopic and non-isotopic internal standards. Analytica Chimica
Acta. 2013;772: 59-67. doi:
10.1016/j.aca.2013.02.013
46.
Broeckling CD, Heuberger AL, Prince JA, Ingelsson E, Prenni JE. Assigning
precursor-product ion relationships in indiscriminant MS/MS data from non-targeted metabolite profiling
studies. Metabolomics. 2013;9: 33-43. doi:
10.1007/s11306-012-0426-4
47.
Wang M, Heo G-Y, Omarova S, Pikuleva IA, Turko IV. Sample Prefractionation
for Mass Spectrometry Quantification of Low-Abundance Membrane Proteins. Anal Chem. 2012;84: 5186-5191. doi:
10.1021/ac300587v
48.
Wang M, Chen J, Turko IV. 15N-Labeled Full-Length Apolipoprotein E4 as an
Internal Standard for Mass Spectrometry Quantification of Apolipoprotein E Isoforms. Anal Chem. 2012;84:
8340-8344. doi:
10.1021/ac3018873
49.
Hoh E, Dodder NG, Lehotay SJ, Pangallo KC, Reddy CM, Maruya KA. Nontargeted
comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry method and software for
inventorying persistent and bioaccumulative contaminants in marine environments. Environ Sci Technol. 2012;46:
8001-8008. doi:
10.1021/es301139q
50.
Lowenthal MS, Gasca-Aragon H, Schiel JE, Dodder NG, Bunk DM. A quantitative
LC-MS/MS method for comparative analysis of capture-antibody affinity toward protein antigens. J Chromatogr B
Analyt Technol Biomed Life Sci. 2011;879: 2726-2732. doi:
10.1016/j.jchromb.2011.07.037
51.
Charvet C, Liao W-L, Heo G-Y, Laird J, Salomon RG, Turko IV, et al.
Isolevuglandins and Mitochondrial Enzymes in the Retina. J Biol Chem. 2011;286: 20413-20422. doi:
10.1074/jbc.M111.232546



